Research and projects
Mission Statement
Our lab is motivated to understand the genetic and cellular mechanisms controlling organ development and to use this knowledge to determine how congenital organ anomalies occur. We use a combination of animal embryo and stem cell-based approaches to recreate the complexity of human development.
We foster a collaborative, inclusive, and growth-oriented environment to achieve our scientific goals and mentor the next generation of scientists.
Congenital organ anomalies are the leading cause of infant mortality in North America.
The disease mechanisms responsible for causing congenital organ anomalies are poorly understood. Though many are caused by known risk genes discovered after genetic sequencing of an individual with a congenital organ anomaly, how sequence variants in these genes affect the underlying cell and developmental biology during organogenesis is not known.
Our lab is particularly interested in how disruptions to cell and tissue fusion-separation events in organogenesis underlie many common congenital malformations. We currently focus on tissue splitting events in the embryonic gut tube, like how the trachea and esophagus split from a single anterior foregut tube.
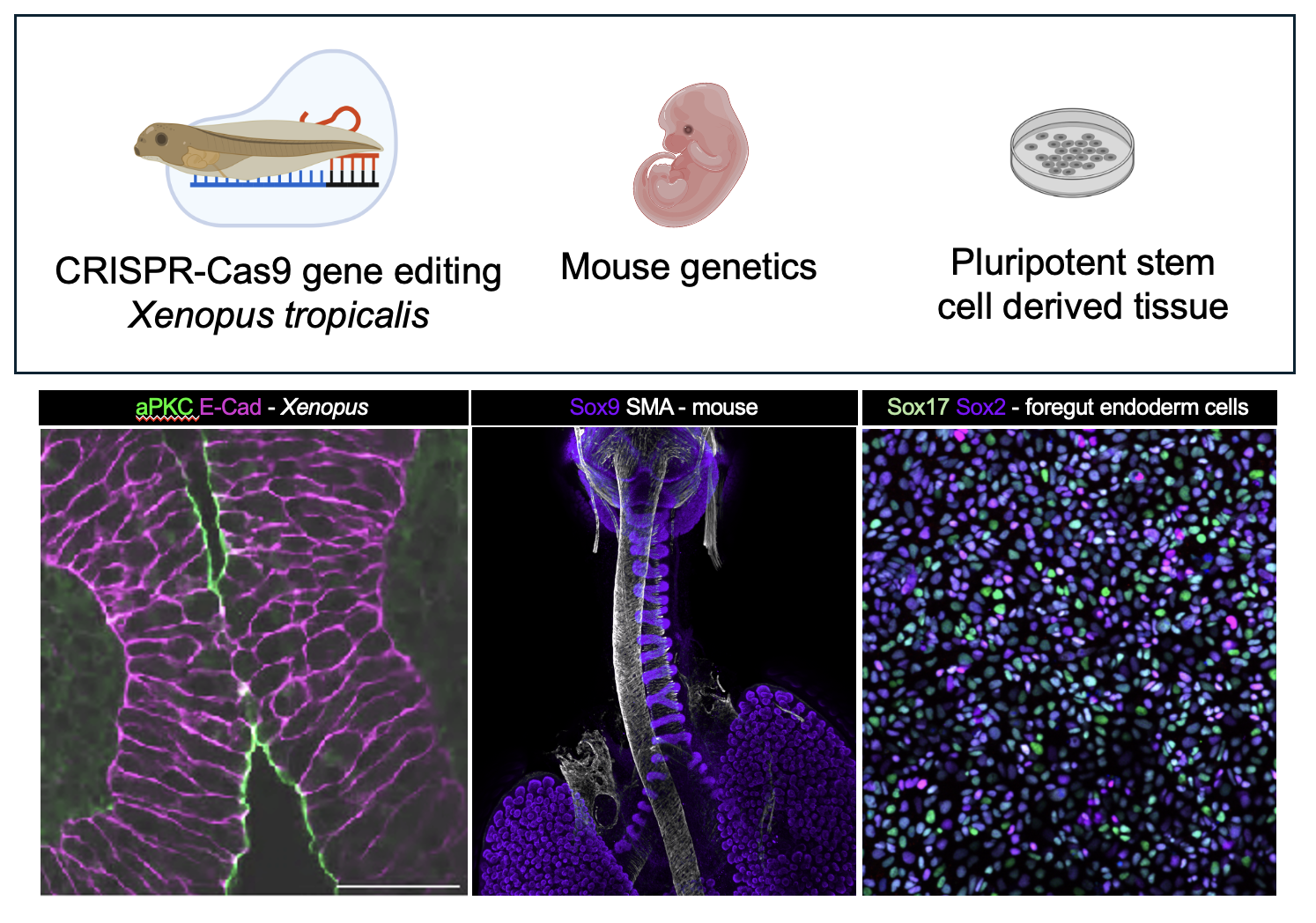
To approach this challenge, our lab uses research animal models including Xenopus (African clawed frog) and mice. Xenopus and mice have all of the same organs as humans, including a trachea and esophagus, and their organ development is similar, making them excellent models to study human development and disease.
We also use pluripotent stem cell-derived digestive and respiratory tissue to understand how the underlying cell biology is disrupted.
Current projects: Endosome trafficking biology during tissue fusion-fission events in the embryonic gut tube
From our human genetics studies we discovered that many patients with trachea and esophageal malformations have genetic variants in proteins involved in endosome trafficking pathways, a pathway all cells use to move proteins to different cellular domains and change the composition of their plasma membrane. We found that endosomes transport polarity proteins to the right place at the right time to keep the trachea and esophagus cells organized during complex tissue fusion and tissue separation events. See our published work here.
Our lab continues to investigate the cell biology controlled by endosome trafficking, identify the upstream signals, and understand how organ malformations that co-manifest with trachea-esophageal anomalies are impacted by disruptions to trafficking pathways.
Long term projects: Developmental bottlenecks in organogenesis
Our long term research goal is to understand how certain organs and tissues are sensitive to loss of critical gene function, particularly in ubiquitously expressed genes and pathways, and how an embryo may overcome adverse developmental phenotypes (gastrulation failure) but later manifest an organ-specific phenotype.
Publications
For a complete list of publications by Nicole Edwards: Google Scholar
Edwards NA, Rankin SA, Kashyap A, Warren A, Agricola Z, Kenny AP, Kofron MJ, Shen Y, Chung WK, Zorn AM. Disrupted endosomal trafficking of the Vangl-Celsr polarity complex underlies congenital anomalies in Xenopus trachea-esophageal morphogenesis. Developmental Cell. 60(18): 2487-2505, 9/2025. PMID: 40412385.
Zhong G*, Ahimaz PR*, Edwards NA*, Hagen JJ, Faure C, Kingma P, Middlesworth W, Khlevner J, El Fiky M, Schindel D, Fialkowski E, Kashyap A, Forlenza S, Kenny AP, Zorn AM, Shen Y, Chung WK. Identification and validation of novel candidate risk genes in endocytic vesicular trafficking associated with esophageal atresia and tracheoesophageal fistulas. Human Genetics and Genomics Advances. 3(3):100107. PMID: 35519826. *Co-first authors
Edwards NA, Shacham-Silverberg V, Weitz L, Kingma PS, Shen Y, Wells JM, Chung WK, Zorn AM. Developmental basis of trachea-esophageal birth defects. Developmental Biology. 477:85-97, 9/2021. PMID: 34023332.